ICP Chain Configuration
Contents
-
ICP Chain Configuration
- Modular Design
-
Available Modules
-
Data-Points Filters
- BoundingBoxDataPointsFilter
- FixStepSamplingDataPointsFilter
- MaxDensityDataPointsFilter
- MaxDistDataPointsFilter
- MaxPointCountDataPointsFilter
- MaxQuantileOnAxisDataPointsFilter
- MinDistDataPointsFilter
- ObservationDirectionDataPointsFilter
- OrientNormalsDataPointsFilter
- RandomSamplingDataPointsFilter
- RemoveNaNDataPointsFilter
- SamplingSurfaceNormalDataPointsFilter
- ShadowDataPointsFilter
- SimpleSensorNoiseDataPointsFilter
- SurfaceNormalDataPointsFilter
- Matchers
- Outlier Filters
- Error Minimizers
- Transformation Checkers
- Inspectors
- Bibliography
-
Data-Points Filters
Modular Design
The registration chain performs a generalization of the Iterative Closest Point algorithm, based on the libpointmachter library. In this chain, a point cloud is a pair of vectors. The first vector is the feature vector, and typically holds the coordinates of the points. The second vector is the descriptor vector, and holds additional informations such as the normals to the points, their colors, etc. The descriptor vector is either empty or of the same size of the feature vector.
The registration chain takes two point clouds as input, the reading and the reference. The registration algorithm tries to align the reading onto the reference. To do so, it first applies some filtering to the cloud, and then iterates. In each iteration, it associates points in reading to points in reference and finds a transformation of reading such as to minimize an alignment error. The following sketch illustrates the process:
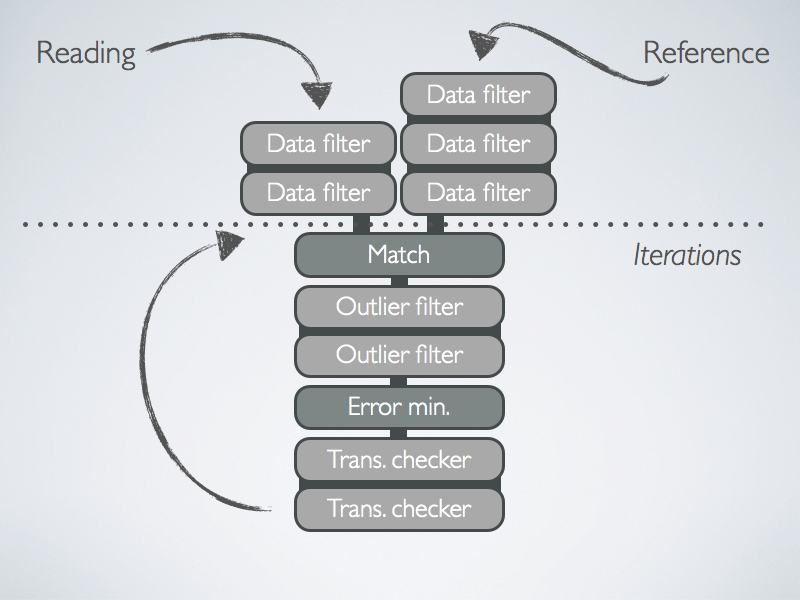
Data-Points Filters
A data-points filter takes a point cloud as input, transforms it, and produces another point cloud as output. For instance, the transformation might add descriptors, such as SurfaceNormalDataPointsFilter, or change the number of points, such as FixStepSamplingDataPointsFilter, or do both, such as SamplingSurfaceNormalDataPointsFilter.
Data-points filters can be chained.
Matchers
A matcher links points in the reading to points in the reference, for now, we provide KDTreeMatcher, based on the fast nearset-neighbor library libnabo.
Feature Outlier Filters
A feature outlier filter removes links between points in reading and their matched points in reference, depending on some criteria. For example, if the distance between the points exceeds a threshold, as in MaxDistOutlierFilter, or if it exceeds a certain number of times the median distance, as in MedianDistOutlierFilter.
Points with no link will be ignored in the subsequent minimization step. Feature outlier filters can be chained.
Error Minimizers
An error minimizer will compute a transformation matrix such as to minimize the error between the reading and the reference. There are different error functions available, such as PointToPointErrorMinimizer or PointToPlaneErrorMinimizer.
Transformation Checkers
A transformation checker can stop the iteration depending on some conditions. For example, a condition can be the number of times the loop was executed, as in CounterTransformationChecker, or it can be related to the matching error, as in DifferentialTransformationChecker.
Transformation checkers can be chained.
Inspector
The inspector allows to log data at the different steps, for analysis.
Available Modules
You configure the ICP chain through a yaml file. You can see an example for openni (launch/openni/icp.yaml of package ethzasl_icp_mapper). The rest of this section shows all available modules and their options. Note that you can retrieve this list using the pmicp -l command of package ethzasl_icp_mapper.
Note that some configurations are invalid, for instance, the PointToPlaneErrorMinimizer depends on the reference cloud having normals in its descriptors. Thus, to use this error minimizer, you must put SurfaceNormalDataPointsFilter or SamplingSurfaceNormalDataPointsFilter as data-point filters to the reference. If the ICP encounters an invalid chain, it will display an error message and abort.
Data-Points Filters
BoundingBoxDataPointsFilter
Subsampling. Remove points laying in a bounding box which is axis aligned.
xMin (default: -1, min: -inf, max: inf)
- minimum value on x-axis defining one side of the bounding box
xMax (default: 1, min: -inf, max: inf)
- maximum value on x-axis defining one side of the bounding box
yMin (default: -1, min: -inf, max: inf)
- minimum value on y-axis defining one side of the bounding box
yMax (default: 1, min: -inf, max: inf)
- maximum value on y-axis defining one side of the bounding box
zMin (default: -1, min: -inf, max: inf)
- minimum value on z-axis defining one side of the bounding box
zMax (default: 1, min: -inf, max: inf)
- maximum value on z-axis defining one side of the bounding box
removeInside (default: 1, min: 0, max: 1)
- If set to true (1), remove points inside the bounding box; else (0), remove points outside the bounding box
FixStepSamplingDataPointsFilter
Subsampling. This filter reduces the size of the point cloud by only keeping one point over step ones; with step varying in time from startStep to endStep, each iteration getting multiplied by stepMult. If use as prefilter (i.e. before the iterations), only startStep is used.
startStep (default: 10, min: 1, max: 2147483647)
- initial number of point to skip (initial decimation factor)
endStep (default: 10, min: 1, max: 2147483647)
- maximal or minimal number of points to skip (final decimation factor)
stepMult (default: 1, min: 0.0000001, max: inf)
- multiplication factor to compute the new decimation factor for each iteration
MaxDensityDataPointsFilter
Subsampling. Reduce the points number by randomly removing points with a density highler than a treshold.
maxDensity (default: 10, min: 0.0000001, max: inf)
- Maximum density of points to target. Unit: number of points per m^3.
MaxDistDataPointsFilter
Subsampling. Filter points beyond a maximum distance measured on a specific axis. If dim is set to -1, points are filtered based on a maximum radius.
dim (default: -1, min: -1, max: 2)
- dimension on which the filter will be applied. x=0, y=1, z=2, radius=-1
maxDist (default: 1, min: -inf, max: inf)
- maximum distance authorized. If dim is set to -1 (radius), the absolute value of minDist will be used. All points beyond that will be filtered.
MaxPointCountDataPointsFilter
Conditional subsampling. This filter reduces the size of the point cloud by randomly dropping points if their number is above maxCount. Based on [1]
prob (default: 0.75, min: 0, max: 1)
- probability to keep a point, one over decimation factor
maxCount (default: 1000, min: 0, max: 2147483647)
- maximum number of points
MaxQuantileOnAxisDataPointsFilter
Subsampling. Filter points beyond a maximum quantile measured on a specific axis.
dim (default: 0, min: 0, max: 2)
- dimension on which the filter will be applied. x=0, y=1, z=2
ratio (default: 0.5, min: 0.0000001, max: 0.9999999)
- maximum quantile authorized. All points beyond that will be filtered.
MinDistDataPointsFilter
Subsampling. Filter points before a minimum distance measured on a specific axis. If dim is set to -1, points are filtered based on a minimum radius.
dim (default: -1, min: -1, max: 2)
- dimension on which the filter will be applied. x=0, y=1, z=2, radius=-1
minDist (default: 1, min: -inf, max: inf)
- minimum value authorized. If dim is set to -1 (radius), the absolute value of minDist will be used. All points before that will be filtered.
ObservationDirectionDataPointsFilter
Observation direction. This filter extracts observation directions (vector from point to sensor), considering a sensor at position (x,y,z).
x (default: 0)
- x-coordinate of sensor
y (default: 0)
- y-coordinate of sensor
z (default: 0)
- z-coordinate of sensor
OrientNormalsDataPointsFilter
Normals. Reorient normals so that they all point in the same direction, with respect to the observation points.
towardCenter (default: 1, min: 0, max: 1)
- If set to true(1), all the normals will point inside the surface (i.e. toward the observation points).
RandomSamplingDataPointsFilter
Subsampling. This filter reduces the size of the point cloud by randomly dropping points. Based on [1]
prob (default: 0.75, min: 0, max: 1)
- probability to keep a point, one over decimation factor
RemoveNaNDataPointsFilter
Remove points having NaN as coordinate.
- no parameters
SamplingSurfaceNormalDataPointsFilter
Subsampling, Normals. This filter decomposes the point-cloud space in boxes, by recursively splitting the cloud through axis-aligned hyperplanes such as to maximize the evenness of the aspect ratio of the box. When the number of points in a box reaches a value knn or lower, the filter computes the center of mass of these points and its normal by taking the eigenvector corresponding to the smallest eigenvalue of all points in the box.
ratio (default: 0.5, min: 0.0000001, max: 0.9999999)
- ratio of points to keep with random subsampling. Matrix (normal, density, etc.) will be associated to all points in the same bin.
knn (default: 7, min: 3, max: 2147483647)
- determined how many points are used to compute the normals. Direct link with the rapidity of the computation (large = fast). Technically, limit over which a box is splitted in two
samplingMethod (default: 0, min: 0, max: 1)
- if set to 0, random subsampling using the parameter ratio. If set to 1, bin subsampling with the resulting number of points being 1/knn.
maxBoxDim (default: inf)
- maximum length of a box above which the box is discarded
averageExistingDescriptors (default: 1)
- whether the filter keep the existing point descriptors and average them or should it drop them
keepNormals (default: 1)
- whether the normals should be added as descriptors to the resulting cloud
keepDensities (default: 0)
- whether the point densities should be added as descriptors to the resulting cloud
keepEigenValues (default: 0)
- whether the eigen values should be added as descriptors to the resulting cloud
keepEigenVectors (default: 0)
- whether the eigen vectors should be added as descriptors to the resulting cloud
ShadowDataPointsFilter
Remove ghost points appearing on edge discontinuties. Assume that the origine of the point cloud is close to where the laser center was. Requires surface normal for every points
eps (default: 0.1, min: 0.0, max: 3.1416)
- Small angle (in rad) around which a normal shoudn't be observable
SimpleSensorNoiseDataPointsFilter
Add a 1D descriptor named <sensorNoise> that would represent the noise radius expressed in meter based on SICK LMS specifications.
sensorType (default: 0, min: 0, max: 2147483647)
- Type of the sensor used. Choices: 0=SickLMS
gain (default: 1, min: 1, max: inf)
- If the point cloud is coming from an untrusty source, you can use the gain to augment the uncertainty
SurfaceNormalDataPointsFilter
Normals. This filter extracts the normal to each point by taking the eigenvector corresponding to the smallest eigenvalue of its nearest neighbors.
knn (default: 5, min: 3, max: 2147483647)
- number of nearest neighbors to consider, including the point itself
epsilon (default: 0, min: 0, max: inf)
- approximation to use for the nearest-neighbor search
keepNormals (default: 1)
- whether the normals should be added as descriptors to the resulting cloud
keepDensities (default: 0)
- whether the point densities should be added as descriptors to the resulting cloud
keepEigenValues (default: 0)
- whether the eigen values should be added as descriptors to the resulting cloud
keepEigenVectors (default: 0)
- whether the eigen vectors should be added as descriptors to the resulting cloud
keepMatchedIds (default: 0)
- whethen the identifiers of matches points should be added as descriptors to the resulting cloud
Matchers
KDTreeMatcher
This matcher matches a point from the reading to its closest neighbors in the reference.
knn (default: 1, min: 1, max: 2147483647)
- number of nearest neighbors to consider it the reference
epsilon (default: 0, min: 0, max: inf)
- approximation to use for the nearest-neighbor search
searchType (default: 1, min: 0, max: 2)
- Nabo search type. 0: brute force, check distance to every point in the data (very slow), 1: kd-tree with linear heap, good for small knn (~up to 30) and 2: kd-tree with tree heap, good for large knn (~from 30)
maxDist (default: inf, min: 0, max: inf)
- maximum distance to consider for neighbors
KDTreeVarDistMatcher
This matcher matches a point from the reading to its closest neighbors in the reference. A maximum search radius per point can be defined.
knn (default: 1, min: 1, max: 2147483647)
- number of nearest neighbors to consider it the reference
epsilon (default: 0, min: 0, max: inf)
- approximation to use for the nearest-neighbor search
searchType (default: 1, min: 0, max: 2)
- Nabo search type. 0: brute force, check distance to every point in the data (very slow), 1: kd-tree with linear heap, good for small knn (~up to 30) and 2: kd-tree with tree heap, good for large knn (~from 30)
maxDistField (default: maxSearchDist)
- descriptor field name used to set a maximum distance to consider for neighbors per point
Outlier Filters
MaxDistOutlierFilter
This filter considers as outlier links whose norms are above a fix threshold.
maxDist (default: 1, min: 0.0000001, max: inf)
- threshold distance
MedianDistOutlierFilter
This filter considers as outlier links whose norms are above the median link norms times a factor. Based on [2].
factor (default: 3, min: 0.0000001, max: inf)
- points farther away factor * median will be considered outliers.
MinDistOutlierFilter
This filter considers as outlier links whose norms are below a threshold.
minDist (default: 1, min: 0.0000001, max: inf)
- threshold distance
SurfaceNormalOutlierFilter
Hard rejection threshold using the angle between the surface normal vector of the reading and the reference. If normal vectors or not in the descriptor for both of the point clouds, does nothing.
maxAngle (default: 1.57, min: 0.0, max: 3.1416)
- Maximum authorised angle between the 2 surface normals (in radian)
TrimmedDistOutlierFilter
Hard rejection threshold using quantile. This filter considers as inlier a certain percentage of the links with the smallest norms. Based on [3].
ratio (default: 0.85, min: 0.0000001, max: 0.9999999)
- percentage to keep
VarTrimmedDistOutlierFilter
Hard rejection threshold using quantile and variable ratio. Based on [4].
minRatio (default: 0.05, min: 0.0000001, max: 1)
- min ratio
maxRatio (default: 0.99, min: 0.0000001, max: 1)
- max ratio
lambda (default: 0.95)
- lambda (part of the term that balance the rmsd: 1/ratio^lambda
Error Minimizers
IdentityErrorMinimizer
Does nothing.
- no parameters
PointToPlaneErrorMinimizer
Point-to-plane error (or point-to-line in 2D). Based on [5]
- no parameters
PointToPointErrorMinimizer
Point-to-point error. Based on SVD decomposition. Based on [6].
- no parameters
Transformation Checkers
BoundTransformationChecker
This checker stops the ICP loop with an exception when the transformation values exceed bounds.
maxRotationNorm (default: 1, min: 0, max: inf)
- rotation bound
maxTranslationNorm (default: 1, min: 0, max: inf)
- translation bound
CounterTransformationChecker
This checker stops the ICP loop after a certain number of iterations.
maxIterationCount (default: 40, min: 0, max: 2147483647)
- maximum number of iterations
DifferentialTransformationChecker
This checker stops the ICP loop when the relative motions (i.e. abs(currentIter - lastIter)) of rotation and translation components are below a fix thresholds. This allows to stop the iteration when the point cloud is stabilized. Smoothing can be applied to avoid oscillations. Inspired by [3].
minDiffRotErr (default: 0.001, min: 0., max: 6.2831854)
- threshold for rotation error (radian)
minDiffTransErr (default: 0.001, min: 0., max: inf)
- threshold for translation error
smoothLength (default: 3, min: 0, max: 2147483647)
- number of iterations over which to average the differencial error
Inspectors
NullInspector
Does nothing.
- no parameters
PerformanceInspector
Keep statistics on performance.
baseFileName (default: )
- base file name for the statistics files (if empty, disabled)
dumpPerfOnExit (default: 0)
- dump performance statistics to stderr on exit
VTKFileInspector
Dump the different steps into VTK files.
baseFileName (default: point-matcher-output)
- base file name for the VTK files
dumpPerfOnExit (default: 0)
- dump performance statistics to stderr on exit
Bibliography
[1] - Registration and integration of multiple range images for 3-D model construction. Masuda, T. and Sakaue, K. and Yokoya, N. In Pattern Recognition, 1996., Proceedings of the 13th International Conference on. 879--883. 1996. DOI: 10.1109/ICPR.1996.546150. full text.
[2] - Simultaneous Localization and Mapping with Active Stereo Vision. Diebel, J. and Reutersward, K. and Thrun, S. and Davis, J. and Gupta, R. In Intelligent Robots and Systems, 2004. (IROS 2004). Proceedings. 2004 IEEE/RSJ International Conference on. 3436--3443. 2004. DOI: 10.1109/IROS.2004.1389948. full text.
[3] - The Trimmed Iterative Closest Point Algorithm. Chetverikov, D. and Svirko, D. and Stepanov, D. and Krsek, P. In Pattern Recognition, 2002. Proceedings. 16th International Conference on. 545--548. 2002. DOI: 10.1109/ICPR.2002.1047997. full text.
[4] - Outlier robust ICP for minimizing fractional RMSD. Phillips, J.M. and Liu, R. and Tomasi, C. In 3-D Digital Imaging and Modeling, 2007. 3DIM '07. Sixth International Conference on. 427--434. 2007. DOI: 10.1109/3DIM.2007.39. full text.
[5] - Object modeling by registration of multiple range images. Chen, Y. and Medioni, G. In Robotics and Automation, 1991. Proceedings., 1991 IEEE International Conference on. 2724--2729. 1991. DOI: 10.1109/ROBOT.1991.132043. full text.
[6] - A Method for Registration of 3-D Shapes. Besl, P.J. and McKay, H.D. In Pattern Analysis and Machine Intelligence, IEEE Transactions. 239--256. 1992. DOI: 10.1109/34.121791. full text.







